MAIN FEATURES
All these features have been extensively used and tested during my PhD
Full extensibility
- Every module can be extended easily : Score definition, Genes and genomes, genome selection
population filter, genome comparison, etc... - High usage of polymorphism to allow specialization and generic usage
- XML grammar adapted to this extensibility feature for the structure editor
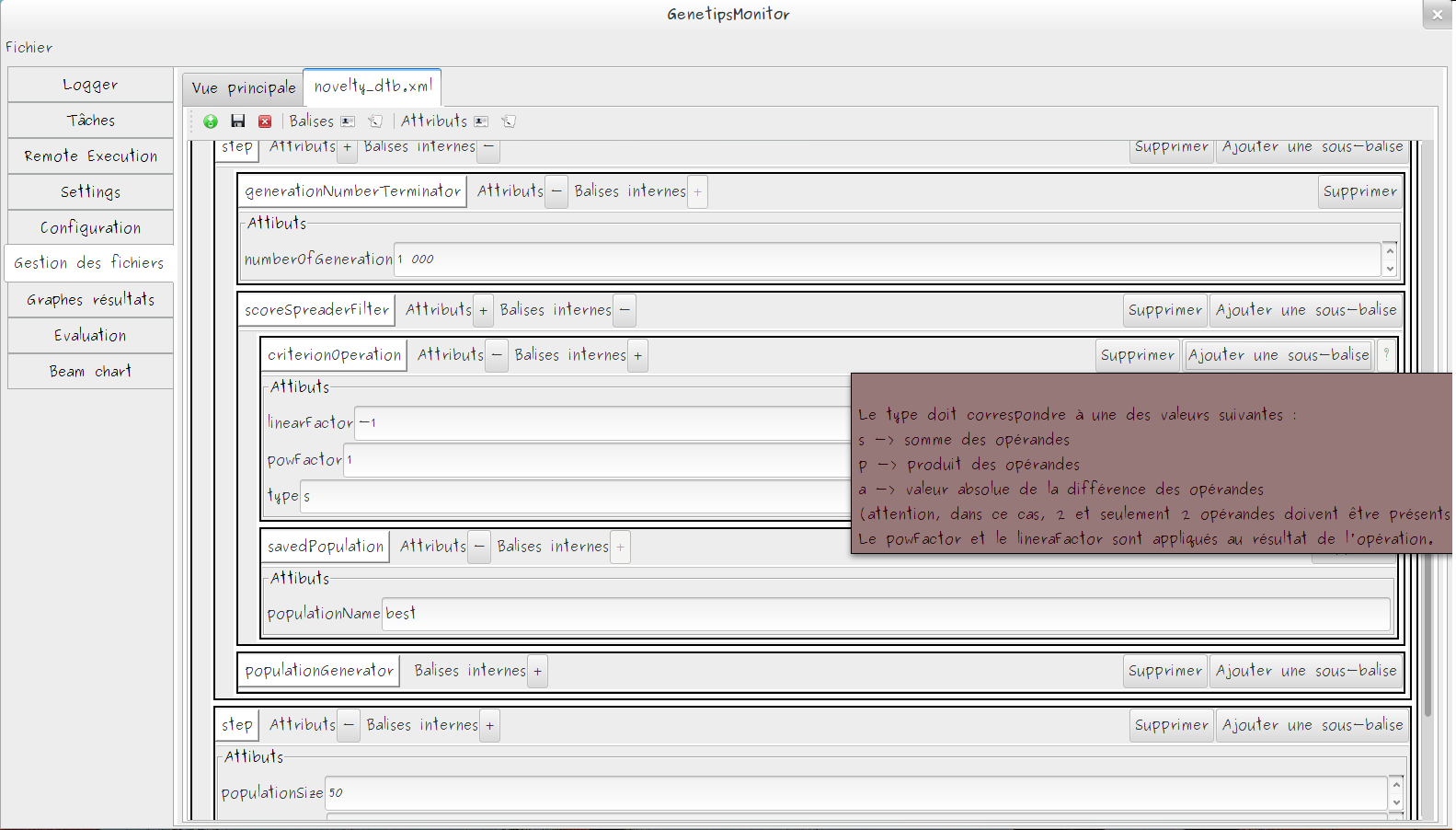
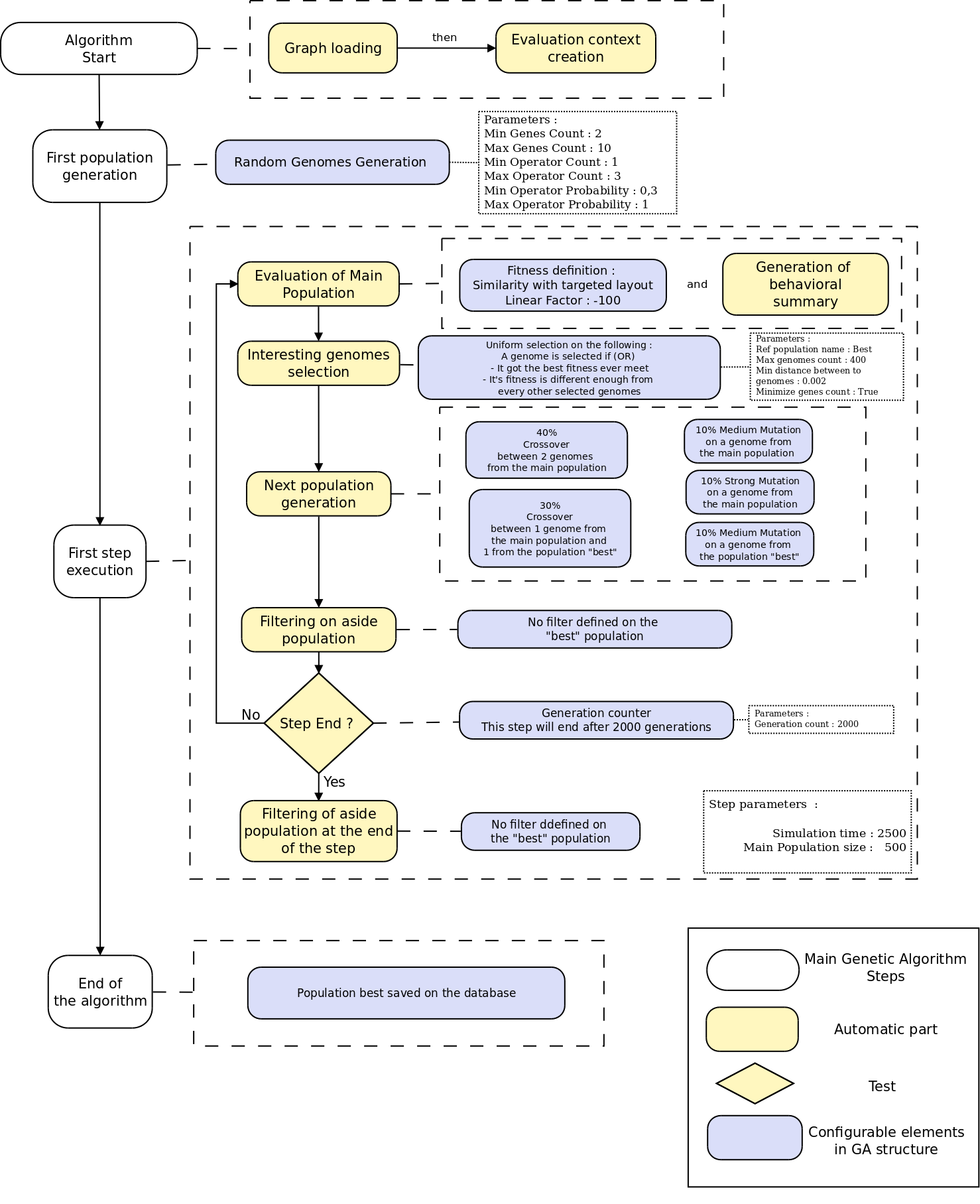
GA Structure Editor
- XML Based editor
- Every block are documented in GUI
- System to automatically discover inheritance link between modules
- Easy to extends block list with a simple block description grammar
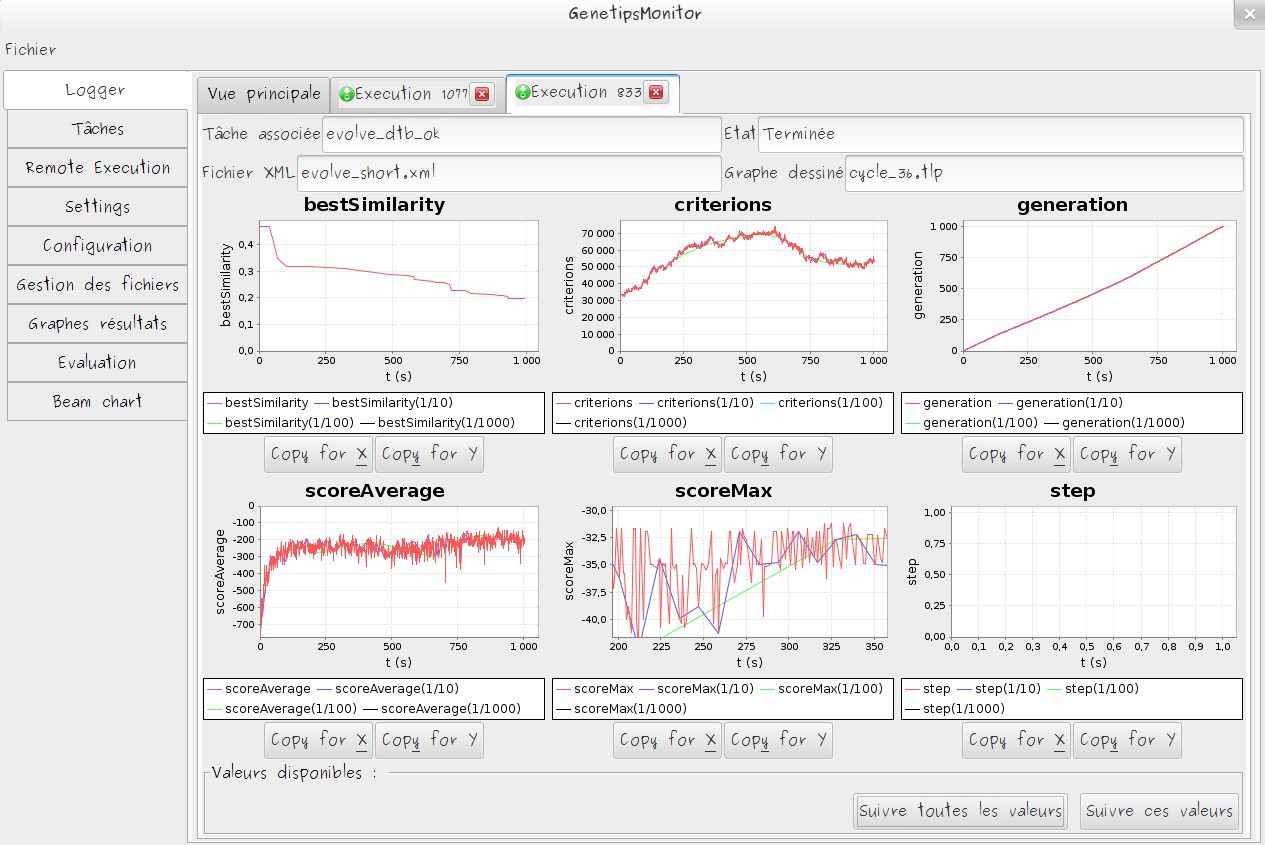
Monitoring
- Automatic monitoring of selected metrics
- Monitoring data are stored server side and could be consulted asynchronously
- DataViz tools to efficiently summarize results of execution set
- Usecase adapted visualization can be easily added
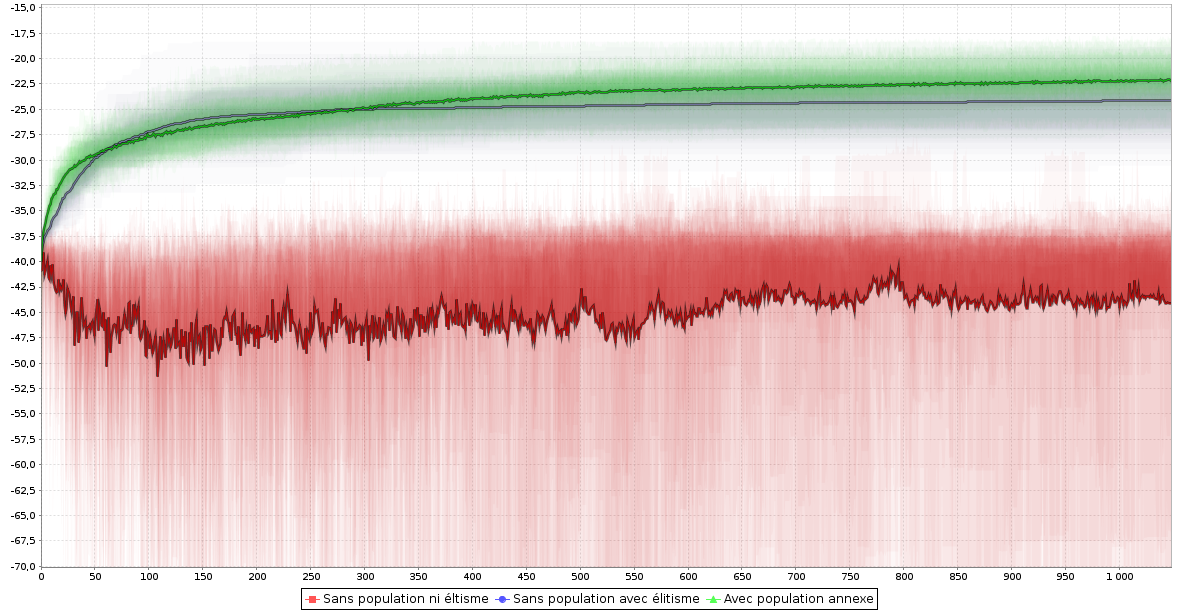
Curiosity
- Curiosity driven fitness definitions
- Interesting genomes are those which behave differently
- Database sample used as reference population
- Allow quick and interesting exploration of the solution space
Roadmap
- Lots of idea for future features
- Selection rule for crossover contained the genome
- Live evolution (not by generation but like in a real enviromnent)
- Local crossover (for 2d or 3d space based usecases), 2 individuals have to be at the same place to generate new genomes by crossover.
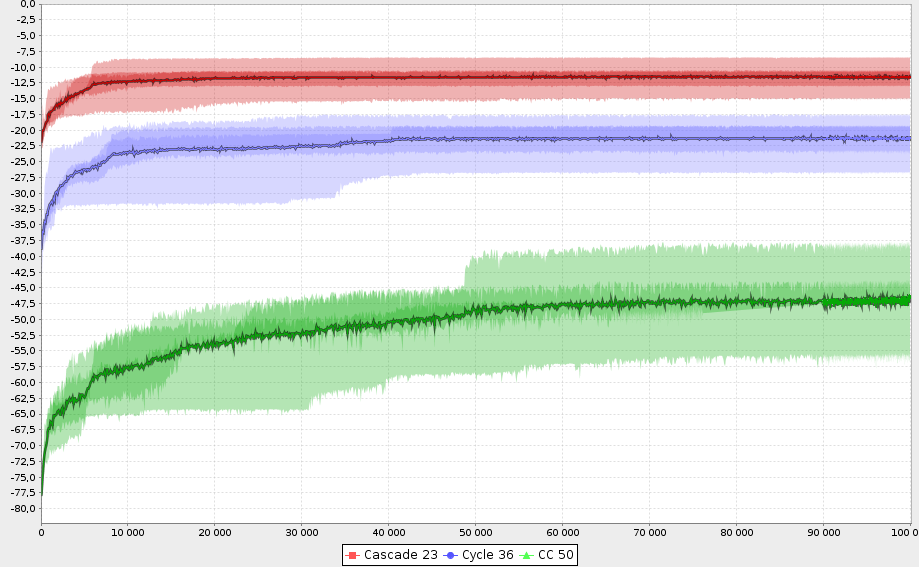
Database
- Heart and memory of GenLib
- Any interesting genome is stored in the database, with evaluation context data and behavior summary
- This allow to bootstrap further executions
- Aside population can be initialized with any SQL query.